Data
Expression tools use TPM (Transcripts Per Kilobase Million) values for absolute expression, and no unit for the Row scaled or relative values. Absolute expression values were generated by aligning RNA-Seq reads to the reference genome and gene annotation with aligned read numbers then used to calculate TPM values.
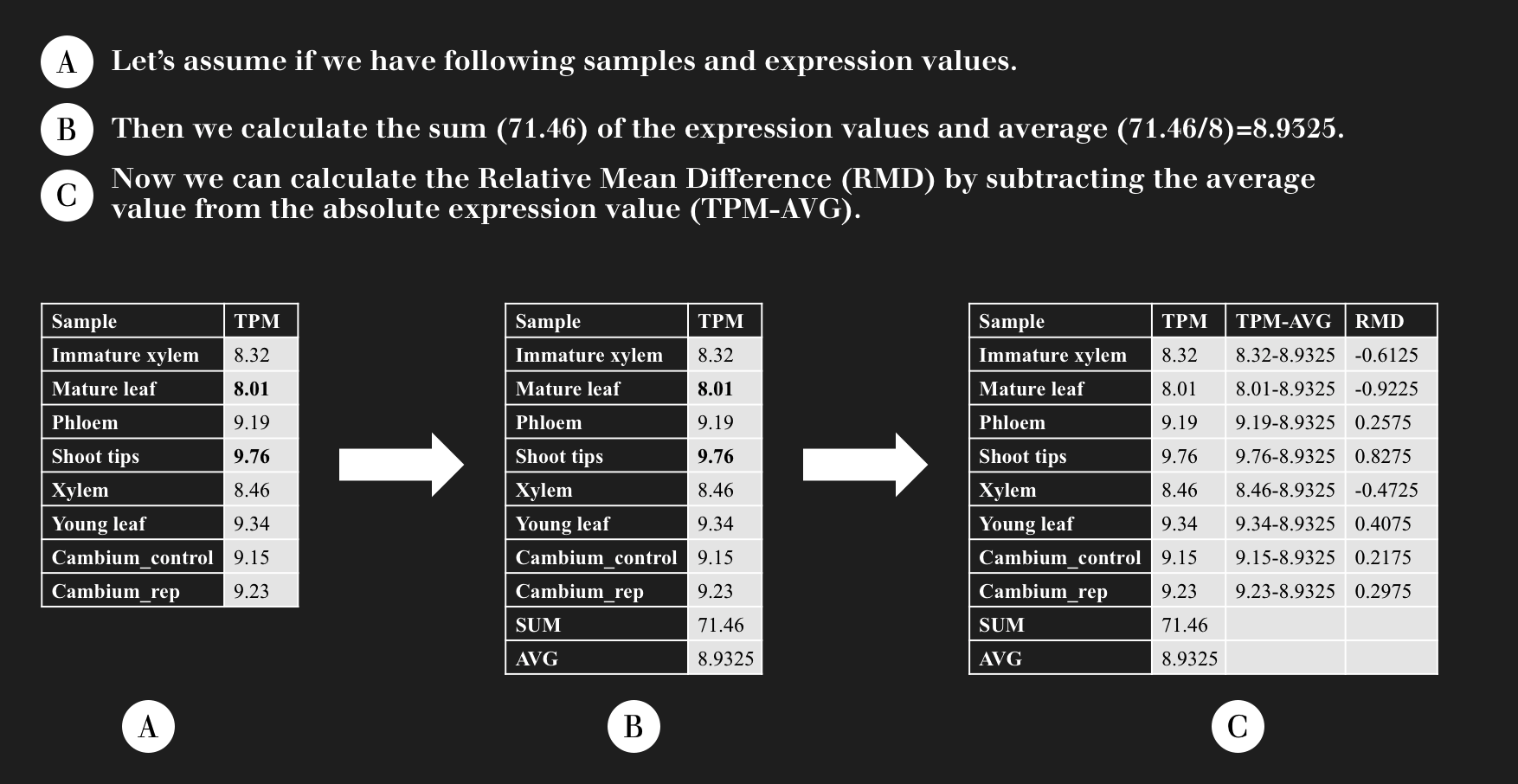
How we calculate the Relative Mean Difference (RMD)?
Gene family construction settings
TribeMCL
Source: Official website
- mclblastline
- I = 2
- scheme = 4
- other parameters are left on their default values
Phylogeny construction settings