When you first go to the https://plantgenie.org, select your species that you will be working with.
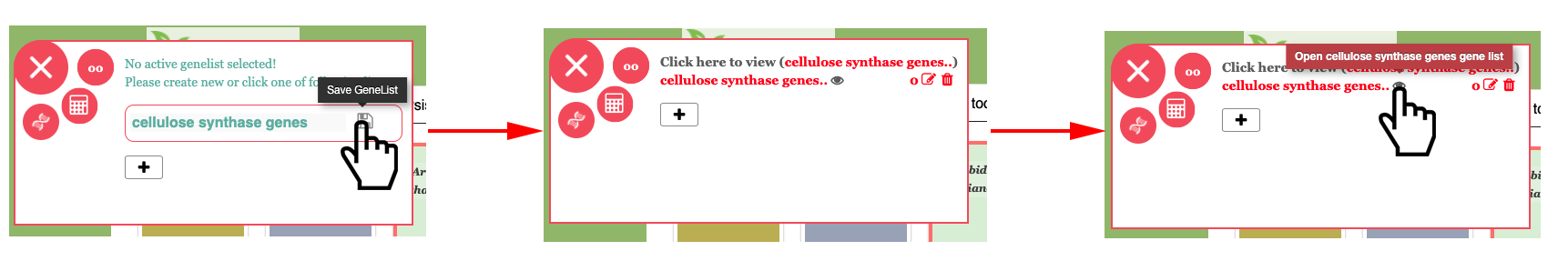
We have selected P. tremula v2.2 as our active species in the above screenshot. Now we are going to start the analysis by creating a gene list. In this example we will be using “cellulose synthase” genes in P. tremula v2.2. In PlantGenIE, first step is always to select the correct species and then create a gene list using the GeneList tool. To create the gene list, click on the floating menu on the left of the site and click the plus to create a new gene list. Enter a name (“cellulose synthase genes.”) and click save. Then open the GeneList tool by clicking on the eye icon or name of your new list as shown in the following figure.
It will take some seconds for the GeneList tool to load and you should wait until you see the list of gene IDs appear before you enter anything into the text search box. Next type in cellulose synthase inside search text input box. Matching results will be shown below. Similarly you can search for gene ID, Description, synonyms or GO IDs inside the search input text box.
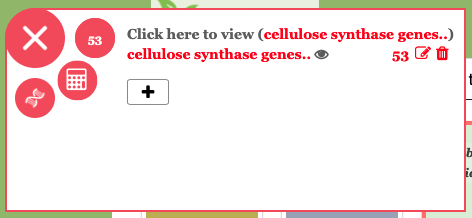
Keep in mind one gene can have multiple splice variants. Therefore we will get different number of entries when we click Show all transcripts and Show all genes buttons. However we only save number of genes with the gene list. Now click on the Save icon (towards the top right) to save cellulose synthase genes to your gene list. Please close the GeneList tool and you will see floating number has been updated with newly created cellulose synthase genes. Finally close the floating menu and start PlantGenIE analysis by navigating to the analysis tools.